Introduction
Our understanding of 'immunome" and its role in the pathogenesis and clinical outcomes of hematological diseases has significantly improved in recent years, which highlights the importance of multidimensional cytometry techniques in investigating immune response both in clinical and research settings. Liquid mass cytometry (LMC), imaging mass cytometry (IMC) and flow cytometry (FC) are powerful techniques for immunophenotyping, biomarker discovery, and patients' immune-monitoring. These techniques provide the ability to profile over 40 markers per cell, resulting in large amounts of data which could be challenging to analyze and interpret. Therefore, we developed ImmunoCluster, an easy-to-use open-source computational pipeline, to explore high dimensionality single-cell cytometry datasets.
Case studies
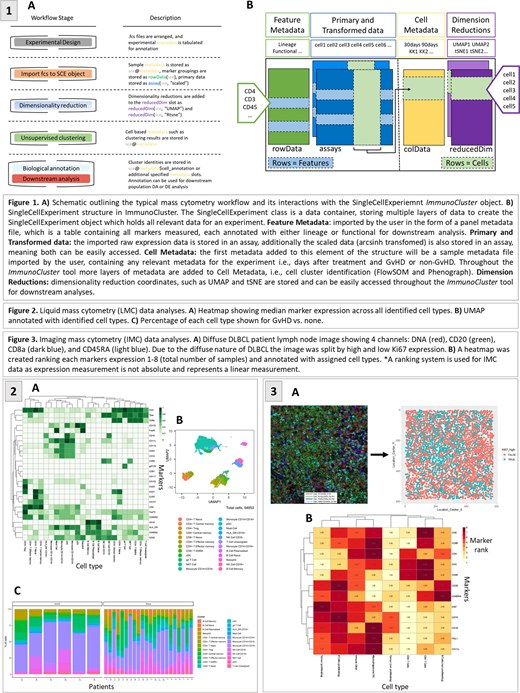
Here we describe three examples of the implementation of the ImmunoCluster tool, which is a self-contained R package (accessed via GitHub: https://github.com/kordastilab/ImmunoCluster). Previously published LMC data from 15 leukemia patients 30 and 90 days after bone marrow transplantation (BMT) were used to test ImmunoCluster's ability to reproduce results. Post-BMT 3/15 patients suffered acute graft versus host disease (GvHD). For IMC data a lymph node section from a diffuse large B-cell lymphoma (DLBCL) patient. Finally, FC data from BM of 7 healthy donors (HDs) taken during hip surgery. The pipeline provides tools to allow researchers to follow a workflow which guides them through experimental design, data analyses and interpretation, to publishable graphics identifying differences in phenotype and abundance of cells between conditions (Figure 1A). ImmunoCluster comprises of three core computational stages:
Stage 1. Data import and quality control
In the experimental design stage, the high dimensional dataset is imported into ImmunoCluster with an associated metadata file, this included timepoints, and response to treatment. All data was stored within a SingleCellExperiment (SCE) object, a data container in which you can store/retrieve information such as metadata and UMAP/tSNE coordinates (Figure 1B). Initial exploratory visualization of the data, e.g. Multidimensional scaling (MDS) plots, and heatmaps showing marker expression for each patient were created and metadata used for annotation.
Stage 2. Dimensionality reduction and unsupervised clustering
We used three dimensionality reduction tools: MDS, uniform manifold approximation and projection (UMAP), and t-Distributed Stochastic Neighbor Embedding (tSNE). Two clustering algorithms: an ensemble clustering method of FlowSOM and Consensus clustering; and PhenoGraph. The aim of these algorithms were to assign all cells to clusters corresponding to true cell types.
Stage 3. Annotation and differential testing
Tools exploring cluster marker expression via projection onto UMAP/tSNE, or heatmaps, aided the identification of cell types and phenotypically distinct clusters. Metadata were used to annotate figures, allowing for visualization of the distribution of cell islands between different conditions/timepoints. Tools such as median marker expression, hierarchical clustered heatmaps, and box plots of cell cluster abundance were applied. Statistically significant differences between conditions were identified.
Results
We successfully replicated the findings from Hartmann et al., 24 cell populations were identified (Figure 2A-B). Significant differences between memory B-cells (FDR p=4.38 x 10-3), naïve B-cells (FDR p=1.35 x 10-2), and naïve CD4+ T-cells (FDR p=3.47 x 10-2) were identified between the GvHD and none (Figure 2C). From the FC HD CD4+ BM population data we were able to identify regulatory T cells (Tregs), including subpopulations, Treg A and Treg B (as low as 0.7% (0.1-2.0) and 0.9% (0.2-2.4), respectively). A marker expression ranking tool was applied to the DLBCL patient IMC data. We identified the majority of Ki-67 high population were proliferating tumor cells (84%), and the Ki-67 low population consisted of a heterogeneous collection of immune cell populations (Figure 3). These case studies show that ImmunoCluster could help clinicians and researchers with varying experience in computational biology to drive their projects from experimental design, wet lab/clinical trial, through to the data analysis process and visualization.
McLornan:JAZZ PHARMA: Honoraria, Speakers Bureau; NOVARTIS: Honoraria, Speakers Bureau; CELGENE: Honoraria, Speakers Bureau. Harrison:Gilead Sciences: Honoraria, Speakers Bureau; Incyte Corporation: Speakers Bureau; Janssen: Speakers Bureau; Sierra Oncology: Honoraria; Novartis: Honoraria, Research Funding, Speakers Bureau; AOP Orphan Pharmaceuticals: Honoraria; Shire: Honoraria, Speakers Bureau; Promedior: Honoraria; Roche: Honoraria; Celgene: Honoraria, Research Funding, Speakers Bureau; CTI Biopharma Corp: Honoraria, Speakers Bureau. Kordasti:Celgene: Research Funding; Novartis: Research Funding; Alexion: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal